|
Protocol
for RNA Quantitation using Ethidium Bromide (etbr)
Robert
W. Fisher, Department of Radiation Oncology and the
Curriculum in Toxicology, University of North Carolina
at Chapel Hill, USA.
1.
Introduction
Detection
and quantitation of nucleic acids is an essential
technique in the molecular biology laboratory. Nucleic
acids are commonly stained with fluorescent dyes when
separated on the basis of size in agarose gels. These
fluorescent dyes are also used to quantitate nucleic
acids in solution. The high sensitivity and selectivity
of fluorometric assays over spectrophotometric methods
has led to the development of instruments designed
to quantitate nucleic acids based on their interactions
with fluorescent dyes. There are a multitude of fluorescent
nucleic acid dyes such as ethidium bromide, propidium
iodide, SYBR™ Green I and II, PicoGreen™
, and Hoechst 33258 Dye. Some of the newer dyes offer
sensitivities in the picogram range. These fluorochromes
allow the quantitation of DNA in the presence of contaminating
protein. The most sensitive dyes are often specific
for double-stranded nucleic acids over single-stranded
nucleic acids. This specificity is useful if there
is a high level of single-stranded RNA present to
interfere with the measurement. On the other hand,
the need to quantitate RNA often arises in the molecular
biology laboratory and the selectivity of the DNA
specific dyes becomes a severe handicap.
One
way around this problem is to use a dye that binds
to nucleic acids in general, whether they be single-
or double-stranded. Probably the most common dye used
for this purpose is ethidium bromide. Ethidium bromide
is a polycyclic fluorescent dye that binds to double-stranded
DNA molecules by intercalating a planar group between
the stacked base pairs of the nucleic acid. Ethidium
bromide can also bind to secondary structure in single-stranded
RNA molecules: regions of local base pairing offer
the stacked base pairs necessary for the dye molecules
to intercalate. When excited by light at or near 546
nm, the dye-nucleic acid complex exhibits an increased
(about 20 fold) fluorescent yield at an emission wavelength
of 590 nm.
The
following describes an ethidium bromide based fluorometric
method for quantitation of RNA solutions using a Turner
BioSystems TD-700 Laboratory Fluorometer.
2.
Materials Required
- TD-700
Fluorometer with standard PMT and 10 mm x 10 mm square
cuvette adaptor (P/N 7000-009)
- Clear
Quartz Lamp (P/N 10-046) or Quartz-halogen lamp (P/N
7000-930)
- Excitation
Filter 550 nm (P/N 034-0500)
- Emission
Filter >570 nm (P/N 10-052R)
- 10
mm x 10 mm Glass Square Cuvettes (P/N 7000-955)
- Transfer
RNA (Sigma Chemical)
- DEPC
H2O or Ultrapure H2O
- 10X
TNE Buffer Stock Solution (100 mM Tris, 2 M NaCl,
10 mM EDTA, pH 7.4)
- Ethidium
Bromide (5-10 mg/mL)
3.
Experiment Protocol
Three
series of experiments were designed to:
1.
Test the linearity and reproducibility of RNA measurements
under the conditions used for DNA quantitation using
Hoechst 33258 Dye; 2. Optimize the ethidium bromide
concentration used for RNA quantitation; and 3. Determine
whether a typical laboratory sample of total RNA can
be quantitated.
Fresh
1X TNE buffer was diluted from the stock 10X TNE buffer
daily. Immediately before use, assay buffer was prepared
by adding ethidium bromide to the 1X TNE buffer to
achieve a final concentration of 0.1 ug/mL, 1.0 ug/mL,
or 10 ug/mL. Assay buffer was kept in a light-proof
container during use. In each experiment, 2 mL of
the assay buffer was used to zero the fluorometer.
Two uL of each tRNA sample was added to 2 mL of the
assay buffer, mixed, and inserted into the fluorometer
for measurement.
In
the first three experiments, standard solutions of
transfer RNA (tRNA) were prepared by dissolving tRNA
in DEPC-treated H2O to a final concentration of 3.724,
1.790, or 1.742 mg/mL as verified by measuring the
A260 on a Beckman DU70 Spectrophotometer. In the first
experiment, three separate sets of serial dilutions
were prepared to determine reproducibility. In the
second and third experiments, each one of a set of
serial dilutions was measured in triplicate.
In
the final experiment, total RNA was isolated from
human mammary epithelial cell (HMEC) line 184B5 using
the acid guanidinium thiocyanate procedure of Chomczynski
and Sacci. The precipitated total RNA was dissolved
in DEPC-treated H2O to a final concentration of 1.700
mg/mL as verified on a spectrophotometer. Fluorometric
measurements were made using an ethidium bromide concentration
of 1.0 µg/mL.
All
measurements were made at room temperature on a Turner
BioSystems TD-700 Fluorometer. The instrument was
operated in the multi-optional mode, measuring raw
fluorescence. The sensitivity was auto-set as described
in the operating manual. Standard precautions were
taken to avoid RNase contamination problems.
4.
Results
- Determining
Reproducibility & Linearity: The first experiment
was designed to determine the reproducibility of RNA
measurements and to verify whether a linear concentration
vs. fluorescence response occurs under conditions
similar to those used in measuring DNA samples with
Hoechst 33258 Dye. The 3.724 mg/mL standard tRNA solution
was used to prepare three separate serial dilutions
with final concentrations of 2.000 mg/mL, 1.000 mg/mL,
and 0.500 mg/mL. The final ethidium bromide concentration
in the assay buffer was 1.0 µg/mL. Each serial dilution
was measured once and a linear concentration vs. fluorescence
curve was obtained, yielding an r2 value of 0.983
(Figure 1).
In
order to test linearity at lower RNA concentrations,
tRNA samples from 1.790 µg/mL down to 112 ng/mL
were measured. Each sample was measured in triplicate,
and again the results were highly linear (r2 = 0.999)
(Figure 2).
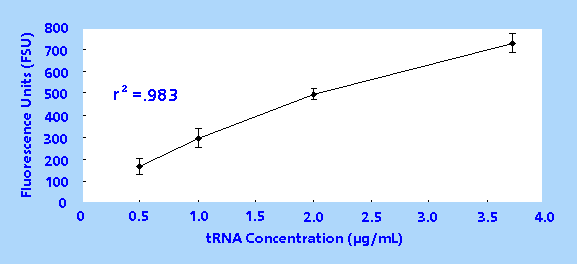
Figure 1. Transfer RNA Concentration vs. Fluorescence
Response
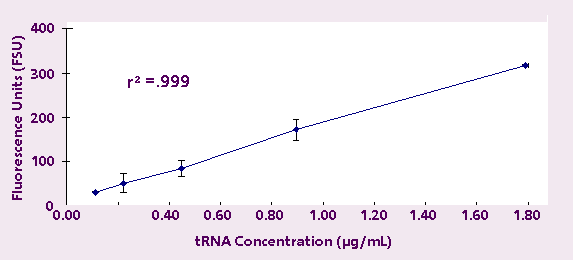
Figure 2. Fluorescence Response of Transfer RNA
down to 112 ng/mL
- Optimizing
Ethidium Bromide Concentration: The next experiment
was designed to determine the optimal concentration
of ethidium bromide. One µg/mL ethidium bromide was
chosen in the preceding experiments because it is
at the high limit of concentrations used to stain
agarose gels for RNA. Final tRNA concentrations of
1.742 µg/mL, 0.871 µg/mL, 0.436 µg/mL, 0.218 µg/mL
and 0.109 µg/mL were measured in triplicate using
10 µg/mL, 1.0 µg/mL and 0.1 µg/mL of ethidium bromide
in the TNE buffer. The results, as shown in Table
1 and Figure 3, indicate an optimal ethidium bromide
concentration of 1.0 µg/ml. Using 1.0 µg/ml of ethidium
bromide one achieves high reproducibility, strong
linear response, and sufficiently low background fluorescence
to measure less than 100 ng/mL RNA.
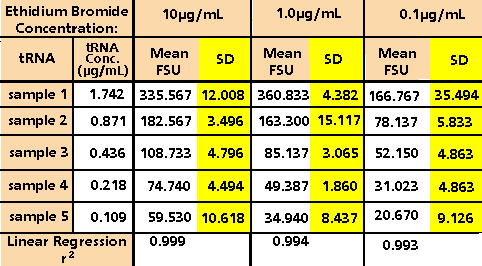
Table 1. Fluorescence of Transfer RNA Using Varying
Concentrations of Ethidium Bromide
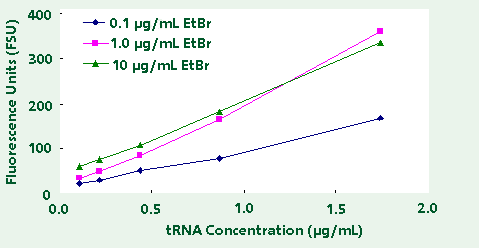
Figure 3. Fluorescence of Transfer RNA using different
EtBr concentrations.
- Measuring
Total RNA: Finally, it was determined that a typical
laboratory sample of total RNA can be quantitated
using the TD-700 Fluorometer. Total RNA from human
mammary epithelial cells was isolated. Total RNA concentrations
of 1.700µg/ml, 0.850µg/ml, 0.425µg/ml and 0.213µg/ml
were measured. Figure 4 shows the fluorescence response
of these samples.
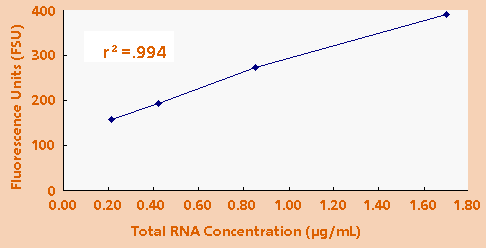
Figure 4. Fluorescence of HMEC total RNA using 1.0µg/mL
EtBr.
5.
Discussion
The
experimental results demonstrate that the Turner BioSystems
TD-700 Fluorometer can quantitate both transfer and
total RNA using ethidium bromide as the fluorochrome.
The procedure is fast, simple, and can be done using
a minimum sample volume. An additional benefit is
that the same TNE buffer can be used to measure DNA
concentrations, changing only the filter sets and
substituting Hoechst 33258 Dye for ethidium bromide
in the assay buffer (See Turner BioSystems Application
Note "A Method for DNA with Hoechst 33258 Dye
and Thiazole Orange").
It
should be noted, however, that a standard curve be
generated using RNA similar to the RNA to be analyzed.
For example, an accurate measurement of total RNA
would not be possible using a standard curve generated
from transfer RNA. On the other hand, if a number
of samples are to be compared to each other (to normalize
gel loading, for example) then a transfer RNA standard
should be adequate.
6.
References
- F.A.
Ausubel, et al., Eds., Current Protocols in Molecular
Biology (John Wiley & Sons, New York, © 1996.
- P.
Chomczynski, N. Sacchi, Anal.Biochem. 162, 156-159,
© 1987.
- J.B.
LePecq, C. Paoletti, Anal. Biochem. 17, 100-107, ©
1966.
- P.N.
Gray, G.F. Saunders, Biochim. Biophys. Acta 254, 60-77,
© 1971.
- M.G.
Murray, H.E. Paaren, Anal.Biochem. 154, 638-642, ©
1986.
- K.
VanDyke, C. Szustkiewicz, Anal.Biochem. 23, 109-115,
© 1968.
Note:
To optimize RNA quantitation, Turner BioSystems tested
various lamp and filter configurations. Using the following,
we were able to maintain linearity down to 50 ng/mL
tRNA:
- Daylight
White Lamp (10-045) with excitation filter 034-0500
and emission filter 10-052
- 10
x 10 mm methacrylate cuvettes (7000-959)
- 3
mL sample volume
|

