|
Protocol
for DNA Quantitation Using PicoGreen®
1.
INTRODUCTION
PicoGreen
dsDNA Quantitation Reagent is an ultra-sensitive fluorescent
nucleic acid stain for quantitating double-stranded
DNA (dsDNA) in molecular biological procedures such
as cDNA synthesis for library production and DNA fragment
purification for subcloning, as well as diagnostic applications,
such as quantitating DNA amplification products1,2
and primer extension assays.3,4
The most commonly used technique for measuring nucleic
acid concentration is the determination of absorbance
at 260 nm (A260) The major disadvantages
of the absorbance method are the large relative contribution
of nucleotides, single-stranded nucleic acids and proteins
to the signal, the interference caused by contaminants
commonly found in nucleic acid preparations, the inability
to distinguish between DNA and RNA, and the relative
insensitivity of the assay (an A260 of 0.1
corresponds to a 5 µg/mL dsDNA solution). Hoechst
(bis-benzimide) dyes are sensitive fluorescent nucleic
acid stains that circumvent many of these problems.
The Hoechst 33258 Dye - based assay is somewhat selective
for dsDNA, does not show significant fluorescence enhancement
in the presence of proteins and allows the detection
and quantitation of DNA concentrations as low as 5.0
ng/mL DNA.5
The Picofluor™ Handheld Fluorometer used
in conjunction with Molecular Probes' PicoGreen dsDNA
Quantitation Reagent enables researchers to quantitate
as little as 1 ng/mL of dsDNA. This sensitivity exceeds
that achieved with the Hoechst 33258 Dye. The standard
PicoGreen assay protocol is also simpler than the Hoechst
33258 method, because a single concentration of the
PicoGreen Reagent allows detection over the full dynamic
range of the assay. In order to obtain the full dynamic
range with Hoechst-based assays, two different dye concentrations
are recommended. In contrast, the linear detection range
of the PicoGreen assay in the Picofluor Fluorometer
extends four orders of magnitude in DNA concentration
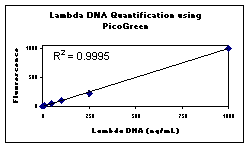
- from 1 ng/mL to 1000 ng/mL with a single dye concentration
(see Figures 1 and 2). This linearity is maintained
in the presence of several compounds commonly found
to contaminate nucleic acid preparations, including
salts, urea, ethanol, chloroform, detergents, proteins
and agarose. The assay protocol has been developed to
minimize the fluorescence contribution of RNA and single-stranded
DNA (ssDNA). Using the PicoGreen dsDNA Quantitation
Reagent and the Picofluor™ Fluorometer,
researchers can quantitate dsDNA in the presence of
equimolar concentrations of ssDNA and RNA with minimal
effect on the quantitation results.
Figure
1 . DNA Standard calibration plot.
2.
MATERIALS REQUIRED
- Picofluor™
Fluorometer with blue optical configuration (P/N 8000-003
or 8000-004)
- 10
mm X 10mm methacrylate fluorescence cuvettes (P/N
7000-959)
- PicoGreen
dsDNA Quantitation Reagent, supplied by Molecular
Probes, Inc., Eugene, Oregon, catalog number P-7581.
A single 1-mL unit of the reagent concentrate is sufficient
for 200 assays using an assay volume of 2 mL and the
protocol described in section 3. Handling, storage
and use of the reagent should be performed in accordance
with the product information sheet supplied by Molecular
Probes, Inc.
3.
EXPERIMENT PROTOCOL
3.1
Reagent Preparation
The PicoGreen dsDNA Quantitation Reagent is supplied
as a 1-mL concentrated dye solution in anhydrous dimethylsulfoxide
(DMSO). On the day of the experiment, prepare an aqueous
working solution of the PicoGreen Reagent by making
a 1:200 dilution of the concentrated DMSO solution
in 10 mM Tris-HCl, 1 mM EDTA, pH 7.5 (TE). To prepare
enough working solution to assay 20 samples, add 100
µL PicoGreen dsDNA Quantitation Reagent to 20.0
mL TE. Preparing this solution in a plastic container
is recommended, as the reagent may adsorb to glass
surfaces. Protect the working solution from light
by covering it with foil or placing it in the dark,
as the PicoGreen Reagent is susceptible to photodegradation.
For best results, this solution should be used
within a few hours of its preparation.
3.2 DNA Standard Curve
3.2.1 Prepare a 2 µg/mL stock solution
of dsDNA in TE. Determine the DNA concentration
on the basis of absorbance at 260 nm (A260) in a
cuvette with a 1-cm pathlength; an A260
of 0.04 corresponds to 2 µg/mL dsDNA solution.
Calf thymus DNA is commonly used for a standard
curve, although any purified dsDNA preparation may
be used. It is preferable to prepare the standard
curve with DNA similar to the type being assayed;
long or short linear DNA fragments for quantitating
similar-sized restriction fragments; plasmid for
quantitating plasmid DNA. However, most linear dsDNA
molecules have been found to yield approximately
equivalent signals, regardless of fragment length.
The PicoGreen assay remains linear in the presence
of several compounds that commonly contaminate nucleic
acid preparations, although the signal intensity
may be affected. Thus, to serve as an effective
control, the dsDNA solution used to prepare the
standard curve should be treated the same way as
the experimental samples and should contain similar
levels of such compounds. To generate a single-replicate,
five-point standard curve from 1 ng/mL to 1000 ng/mL,
proceed to step 3.2.2.
3.2.2 For the DNA standard curve, dilute
the 2 µg/mL DNA stock solution into acrylic
cuvettes as shown in Table 1. Then add 1.0 mL of
the aqueous working solution of PicoGreen Reagent
(prepared in section 3.1) to each cuvette. Mix well
and incubate for 2 to 5 minutes at room temperature,
protected from light.
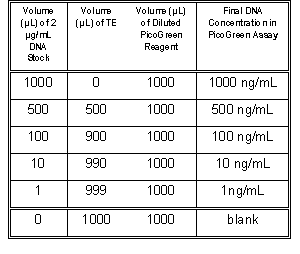
Table 1. Protocol for preparing standard curve.
3.2.3 After incubation, measure the sample
fluorescence in the Picofluor™ Fluorometer.
Press the "CAL" button and then "Enter".
Next, insert blank from chart above and press "Enter".
After blank is read insert the most fluorescent
sample (1 µg/mL DNA) and press "Enter".
After the standard is read, press "Enter".
(The Picofluor flashes calibration values for the
Blank and Standard. The value for the standard should
always be higher than the blank.)
3.2.4
Measure the fluorescence of the remaining samples.
To equalize any photobleaching effects, insert
samples into the fluorometer for approximately equal
time periods. The fluorescence value of the
reagent blank may be subtracted from that of each
sample. Corrected or uncorrected data may be used
to generate a standard curve of fluorescence versus
DNA concentration.
4.
REFERENCES
1.
Nucleic Acids Res. 24, 2623 (1996)
2. BioTechniques 21, 372 (1996)
3. BioTechniques 21, 664 (1996)
4. Proc. Natl. Acad. Sci. USA 93, 6091 (1996)
5. Anal. Biochem. 102, 344 (1980)
6. Molecular Cloning: A Laboratory Manual, Second
Edition, J. Sambrook, E.F. Fritsch and T. Maniatis,
Cold Spring Harbor Laboratory Press, Cold Spring Harbor,
New York (1989).
5.
WARNINGS AND PRECAUTIONS
The
PicoGreen dsDNA Quantitation Reagent is the subject
of patent applications filed by Molecular Probes,
Inc. and is not available for resale or other commercial
uses without a specific agreement from Molecular Probes,
Inc. PicoGreen is a registered trademark of Molecular
Probes, Inc.
|

