|
Protocol
for CyQUANT™ Cell Proliferation Assay
1.
Introduction
The
Turner BioSystems TD-700 Laboratory Fluorometer in
combination with Molecular Probes' CyQUANTTM
Cell Proliferation Assay Kit provides a convenient,
rapid and sensitive procedure for determining the
density of cells in culture. The assay has a linear
detection range extending from 50 or fewer to at least
200,000 cells in 2 mL volumes (Figure 1) and is therefore
ideal for cell proliferation studies as well as for
routine cell counts. The assay is based on dye fluorescence
enhancement upon binding to cellular nucleic acids.
(1) Cells are simply lysed by addition of a buffer
containing the CyQUANT-GR dye; there are no washing
steps, growth medium changes or long incubations.
The resulting fluorescence is proportional to the
number of cells in the sample and is measured directly
using the TD-700 fluorometer equipped with a fluorescein
filter kit. The CyQUANT assay can detect much lower
cell numbers than Neutral Red or methylene blue assays.
(2,3,4) Unlike procedures that rely on the conversion
of tetrazolium dyes to blue formazan (5) products
or 3H thymidine incorporation assays, (6) the CyQUANT
method is rapid and does not rely on cellular metabolic
activity. Thus, cells can be frozen prior to assaying;
time course assays are facile and data obtained from
samples taken at widely different time intervals can
be directly compared. Also, unlike tetrazolium conversion
assays, serum components do not appreciably interfere
with the assay. The CyQUANT assay performs reliably
with widely disparate cell types, including mouse
fibroblasts (NIH 3T3 and CREBAG 2 cells), normal human
umbilical vein endothelial cells (HUVEC, InvitroCyte,
Inc., Seattle), canine kidney cells (MDCK), chinook
salmon embryo cells (CHSE), rat basophilic leukemia
(RBL), rat glioma cells (C6) and mouse myeloma cells
(P3X63A68).
2.
Materials Required
- TD-700
Fluorometer with standard PMT and 10 mm x 10 mm square
cuvette adaptor (P/N 7000-009).
- Blue
Mercury Vapor Lamp (P/N 10-089) or Quartz-halogen
lamp (P/N 7000-930).
- Fluorescein
filter kit (P/N 10-086R).
- 10mm
x 10 mm square methacrylate disposable cuvettes (P/N
7000-959).
- CyQUANT
Cell Proliferation Assay Kit (catalog number C-7026),
supplied by Molecular Probes, Inc., Eugene, OR. The
kit contains 550µL of 400X CyQUANT-GR stock solution
in DMSO, 11 mL of 20X Cell Lysis Buffer, and 100µL
of 100µg/mL lamda -DNA Standard in TE buffer (10 mM
Tris, 1 mM EDTA). The kit contents are sufficient
for 100 assays using 2.0 mL samples in 10 mm x 10
mm cuvettes. Handling, storage and the use of the
reagents should be performed in accordance with the
product information sheet supplied by Molecular Probes,
Inc.
3.
Experiment Protocol
3.1
Reagent Preparation
On
the day of the experiment, dilute the concentrated
Cell Lysis Buffer stock solution 20-fold in distilled
water (2.0 mL will be required for each assay). Just
prior to running the experiment, dilute the CyQUANT-GR
stock solution 400-fold into the 1X Cell Lysis Buffer.
For example, to prepare 20 mL of CyQUANT-GR working
solution (enough for ~10 assays), first make the 1X
Cell Lysis Buffer by mixing 1 mL of the 20X stock
with 19 mL of nuclease-free distilled water; next
add 50µL of the CyQUANT-GR stock solution and mix
thoroughly. We recommend preparing the working solution
in a plastic container, rather than in glass; the
CyQUANT-GR reagent may adsorb to glass surfaces. Protect
the working solution from light by keeping it in an
opaque bottle, covering it with foil or placing it
in the dark to prevent photodegradation of the CyQUANT-GR
dye. For best results, the solution should be used
within a few hours of its preparation.
3.2
Cell-Number Standard Curve
A
reference standard curve can be created for converting
sample fluorescence values into cell numbers. The
cell type used for the standard curve should be the
same as that which is used in the experiment. It is
possible to assay either suspension cells or adherent
cells, however, the latter must first be detached
and suspended by treatment with trypsin. Note that
some adherent cells are sensitive to trypsinization
and some cell lysis might ensue.
3.2.1
Prepare a concentrated cell suspension in medium:
ideally this should be ~1 mL total volume at a density
of about 5 x 105 - 1 x 106
cells/mL. Determine the actual cell density by counting
the cells using a hemacytometer(7) or optical density
measurements.(8)
3.2.2
Centrifuge 1.0 mL of the concentrated cell suspension
for 5 minutes at 200 X g (1500 rpm in a microcentrifuge).
Carefully remove and discard the supernatant without
disturbing the cell pellet, and freeze the cell
pellet at -70EC (note [A]).
3.2.3
Thaw the cell pellet at room temperature, add 1.0
mL of the CyQUANT-GR dye/Cell Lysis Buffer (prepared
in Section 3.1), and resuspend the cells by briefly
vortexing.
3.2.4
In a set of culture tubes, serially dilute the cell
suspension with CyQUANT-GR dye/Cell Lysis Buffer
to obtain a range of 2.0 mL samples containing numbers
of cells from 50 to 200,000. Include a 2.0 mL sample
with no cells as a reagent blank. Incubate for 2-5
minutes at room temperature, protected from light.
3.2.5
Transfer samples to standard acrylic fluorescence
cuvettes and measure the fluorescence using the
TD-700 fluorometer with the fluorescein filter set
(P/N 10-086R). Insert the most fluorescent sample
first and calibrate the instrument sensitivity as
directed in the TD-700 manual (press #2, calibrate).
This procedure automatically optimizes the instrument
sensitivity to match the fluorescence of the sample.
3.2.6
Measure the fluorescence of the remaining samples.
To equalize any photobleaching effects, insert
samples into the fluorometer for approximately equal
time periods. The fluorescence value of the
reagent blank (CyQUANT-GR dye + Cell Lysis Buffer
only) may be subtracted from that of each sample.
Corrected or uncorrected data may be used to generate
a standard curve of fluorescence versus cell number
(for example, see Figure 1). The form of the standard
curve will vary with cell type.
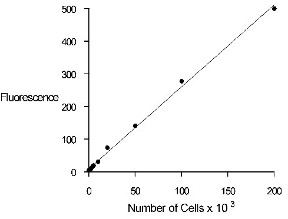
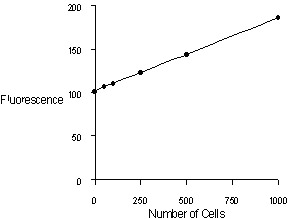
Figure 1. Quantitation of MDCK (Madin-Darby canine
kidney) cells using the CyQUANT Cell Proliferation Assay
Kit and the TD-700 fluorometer. Upper panel: 1000 to
200,000 cells per 2.0 mL sample. Lower panel: 50 to
1000 cells per 2.0 mL sample.
3.3
Cell-Number Determination: Cells Grown in Standard Culture
Conditions
The
CyQUANT Cell Proliferation Assay Kit can be used to
count the number of cells in a sample taken from a
conventional cell culture. Adherent cells must be
first detached by treatment with trypsin and then
suspended. Note that some lysis may occur in adherent
cells that are sensitive to trypsinization. Cells
grown in suspension can be assayed directly.
3.3.1
Transfer suspended cell samples to centrifuge tubes
and centrifuge for 5 minutes at 200 X g (e.g., 1500
rpm in a microcentrifuge). Samples should contain
from about 50 to about 200,000 cells. Remove and discard
the supernatant without disturbing the cell pellet,
and freeze the cell pellet at -70EC (note [A]).
3.3.2
Thaw the cell pellets at room temperature and add
2.0 mL of CyQUANT-GR/Cell Lysis Buffer (prepared in
Section 3.1) to each sample.
3.3.3
Transfer entire 2.0 mL samples to acrylic fluorescence
cuvettes and measure the fluorescence using the TD-700
fluorometer using the same instrument parameters as
used in generating the standard curve (Section 3.2.5).
To equalize any photobleaching effects, insert
samples into the fluorometer for similar time periods
to those used for the standard curve measurements.
3.3.4
If the standard curve was plotted using blank-subtracted
data (Section 3.2.5), the reagent blank (CyQUANT-GR
dye + Cell Lysis Buffer only) fluorescence value must
also be subtracted from that of each of the samples.
Convert the observed fluorescence to cell number using
a standard curve (Section 3.2.6). We have found that
many different cell types can be assayed using this
procedure, but the absolute signal is cell type-dependent.
Thus it is advisable to use a standard curve generated
from the same cell type that is being assayed, for
comparison. Alternatively, a standard curve generated
using pure DNA (Section 3.5) can be calibrated relative
to an appropriate cell type.
3.4
Cell Number Determination Based on DNA or RNA Alone
In
the protocols described above, the CyQUANT-GR reagent
is used to determine cell number by staining both
DNA and RNA. DNA to RNA ratios, however, may vary
according to cell type and cell cycle position. Fluorescence
due to CyQUANT-GR dye binding to RNA can be eliminated
by pretreating samples with DNase-free RNase. Likewise,
fluorescence due to DNA-bound dye can be eliminated
by pretreating samples with RNase-free DNase.
3.4.1
For determination of total cellular DNA or RNA, freeze
a cell pellet containing 20,000-100,000 cells at -70EC,
thaw at room temperature, and resuspend in 1 mL of
1X Cell Lysis Buffer, containing 180 mM NaCl. For
RNase treatment, this buffer should also contain 1
mM EDTA. For DNase treatment the buffer should contain
1 mM CaCl2 and 1 mM MgCl2.
3.4.2
DNase-free-RNase A or RNase-free-DNase I is added
to a final concentration of about 1.35 Kunitz units/mL
(RNase) or 45 Kunitz units/mL (DNase). (9,10) Samples
are incubated for one hour at room temperature.
3.4.3
An equal volume of a 2X working solution of CyQUANT-GR
dye (50µL of the CyQUANT-GR/DMSO stock solution per
10 mL of 1X Cell Lysis Buffer) is added to each sample.
Samples are incubated for 2-5 minutes.
3.4.4
The fluorescence is measured as described above (3.2.5).
It is suggested that controls be run for each digested
sample, using the appropriate buffer, as the presence
of salt and divalent cations slightly reduces the
slope of the standard curve.
3.5
DNA Standard Curve
The
CyQUANT Cell Proliferation Assay Kit includes a 100µg/mL
sample of bacteriophage lambda-DNA that can be used
to prepare a standard curve for DNA content. The standard
curve can serve to quantitate cellular DNA, provided
that the cell lysates are pretreated with DNase-free
RNase to eliminate the RNA component of the fluorescence
signal (Section 3.4, above). Alternatively, the standard
curve can be used to calibrate the assay for use of
the fluorometer at different times or on different
days. Variation in the signal intensity of the standard
curve is directly related to variation that will be
observed for assaying cells on different days, and
is instrument-dependent. In a set of culture tubes,
prepare serially diluted 2.0 mL samples of bacteriophage
lamda-DNA with concentrations ranging from 50 pg/mL
to 0.5µg/mL by diluting the 100µg/mL stock solution
into CyQUANT-GR/Lysis Buffer working solution (as
prepared in Section 3.1). Include a reagent blank
sample without DNA. Measure fluorescence as described
above (3.2.5 and 3.2.6).
Note
[A] Frozen cell pellets in centrifuge tubes can be
stored at -70° C for up to four weeks. The freezing
step is important for efficient cell lysis in the
CyQUANT assay.
4.
References
- J
Immunol Meth 142, 199 © 1991
- In
Vitro Toxicology 3, 219 © 1990
- Biotechnic
and Histochemistry 68, 29 © 1993
- Anal
Biochem 213, 426 © 1993
- Cancer
Res 48, 4827 © 1988
- Exp
Cell Res 124, 329 © 1979
- Meth
Enzymol 58, 141 © 1979
- BioTechniques
21, 260 © 1996
- J
Gen Physiol 24, 15 © 1940
- J
Gen Physiol 33, 349 © 1950
5.
Patent and Trademark Information
CyQUANT
is a trademark of Molecular Probes, Inc. Materials
and methods incorporated in the CyQUANT Cell Proliferation
Assay Kit are covered by current or pending U.S. and
foreign patents.
|

