|
Protocol
for DNA Quanitation using Hoechst 33258 Dye
1.
Introduction
Quantitation
of DNA is a prelude to many practices in Molecular
Biology. Common techniques that use DNA, such as sequencing,
cDNA synthesis and cloning, RNA transcription, transfection,
nucleic acid labeling (e.g. Random prime labeling),
etc., all benefit from a defined template concentration.
Failure to produce results from these techniques can
sometimes be attributed to an incorrect estimate of
the DNA template used.
The
concentration of a nucleic acid is most commonly measured
by UV absorbance at 260 nm (A260). The
average extinction coefficient for double-stranded
DNA (1A260 = 50 µg/mL), single stranded
DNA (1A260 = 33 µg/mL), or RNA (1A260
= 40 µg/mL) is used to quantitate the nucleic acid
directly from the absorbance at this wavelength. For
accurate results, absorbance should be in the range
of 0.05 - 0.10, which for a 1.0 mL assay, requires
2.5 - 5.0 µg of dsDNA. For dilute nucleic acid samples,
the solution to be measured should also be relatively
free of other components that would add significantly
to the absorbance at 260 nm. Because of these limitations,
alternate techniques have been sought that provide
more sensitivity and are less variant to background
absorbance.
One
such alternative for reliable quantitation of DNA
that significantly improves sensitivity and begins
to address the issues of variance is fluorescence.
As with the common practice of visualizing DNA in
a gel with ethidium bromide, quantitation of DNA can
be easily achieved in a fluorometer with the dye,
Hoechst 33258 Dye, a bisbenzimide DNA intercalator
that excites in the near UV (350 nm) and emits in
the blue region (450 nm). Sensitivity of the Hoechst
33258 Dye assay is approximately 1 ng/mL.This dye
overcomes some of the limits associated with quantitation
of dsDNA by absorbance and can be used in the Turner
BioSystems TD-700 Laboratory Fluorometer.
2.
Materials Required
- TD-700
Laboratory Fluorometer with standard PMT (P/N 7000-009)
- Near
UV Lamp (P/N 10-049)
- Excitation
filter (P/N 10-069R)
- Emission
filter (P/N 10-110RC)
- 10mm
x 10mm Methacrylate fluorescence cuvettes (P/N 7000-959)
- Hoechst
33258 Dye stock dye solution
- 10X
TNE buffer stock solution
- 0.45µm
filtered water
3.
Factors to Consider
3.1
The AT% of a DNA sample affects Hoechst 33258 Dye-DNA
fluorescence. Hence, it is important to use a standard
similar to the samples you are testing. Calf Thymus
DNA can often serve as a reference for most plant
and animal DNA because it is double-stranded, highly
polymerized, and is approximately 58%AT (42%GC). For
bacterial DNA, a different standard may be needed
because the AT% varies widely depending on species.
3.2
The conformation (supercoiled, relaxed, circular,
linear) of plasmid DNA may result in different Hoechst
33258 Dye binding efficiencies. Thus, it is important
to select a standard with similar physical characteristics
to your sample. The most stable form would be a linear
one.
3.3
Hoechst 33258 Dye fluoresces only about half as much
when it binds to single-stranded genomic DNA compared
to when it binds to double-stranded genomic DNA. In
addition, short pieces of single-stranded DNA will
not normally cause Hoechst 33258 Dye to fluoresce
in proportion to their concentration.
3.4
Buffers commonly used to extract DNA from whole cells
have little or no effect on this assay.
3.5
Low levels of detergent(less than 0.01% SDS) have
little or no effect on this assay.
3.6
Salt concentrations in the sample extract of up to
3 M NaCl does not affect this assay. For peak fluorescence,
at least 200 mM NaCl is required for purified DNA
and 2.0 to 3.0 M for crude samples. In crude samples,
higher salt concentrations appear to cause the dissociation
of proteins from DNA, allowing the dye molecules to
bind easier to DNA.
3.7
RNA does not interfere significantly with the DNA
assay because Hoechst 33258 Dye does not normally
bind to RNA. Under high salt concentrations, fluorescence
from RNA is usually less than 1% of the signal produced
from the same concentration of DNA.
4.
Solution Preparation
NOTE:
Hoechst 33258 Dye is a possible carcinogen and possible
mutagen. Wear gloves and a mask, and work under a
fume hood.
4.1
Hoechst 33258 Dye stock dye solution (1 mg/ml): Dilute
1 mL Hoechst 33258 Dye(10 mg/mL solution) with 9 ml
Distilled, 0.45 µm filtered water. Store in an amber
bottle at 4°C for up to 6 months.
4.2
10X TNE buffer stock solution: Dissolve into 800 ml
of distilled water:
- 12.11
g Tris base [Tris (hydroxymethyl) aminomethane],
MW = 121.14
- 3.72
g EDTA, disodium salt, dihydrate, MW = 372.20
- 116.89
g Sodium chloride, MW = 58.44
Adjust
pH to 7.4 with concentrated HCl. Add distilled water
to 1000 mL. Filter (0.45 µm) before use. Store at
4°C for up to 3 months.
NOTE:
The pH & NaCl concentration are essential for
the reagent to bind properly.
4.3
Low range assay solution (for 10-500 ng/ml final DNA
concentration): Dilute 10 µL Hoechst 33258 Dye stock
solution (1 mg/mL) with 10 mL 10X TNE and 90 mL Distilled,
0.45 µm filtered water.
NOTE:
Keep assay solution at room temperature. Prepare fresh
daily. Do not filter once dye has been added.
4.4
High range assay solution (for 100-5000ng/ml final
DNA concentration): Dilute 100 µL Hoechst 33258 Dye
stock solution (1 mg/mL) with 10 mL 10X TNE and 90
mL Distilled, 0.45 µm filtered water.
NOTE:
Keep assay solution at room temperature. Prepare fresh
daily. Do not filter once dye has been added.
4.5
1X TNE: Dilute 10 mL 10X TNE with 90 mL Distilled,
0.45 µm filtered water.
4.6
Calf Thymus DNA Standard: Prepare a 1 mg/mL stock
solution of Calf thymus DNA in TE. Gently tap the
tube to mix thoroughly. Store at 4°C for up to 3 months.
5.
Protocol
NOTE:
Accurate pipetting and thorough mixing are critical
for reproducible results. However, take extreme care
when mixing samples; do not introduce air bubbles.
Air bubbles can cause scattering of light leading
to inaccurate results. If air bubbles form, hold the
upper portion of the cuvette in one hand and gently
tap the bottom sides of the cuvette with your other
hand to release bubbles.
5.1
Choose the assay range most suitable for your samples.
If the low range assay(10 to 500 ng/mL final DNA concentration)
is selected, prepare 2 ml of 100 ng/mL DNA by adding
2 µL 100 µg/ml DNA to 2 mL low range assay solution
prepared in 4.3. If the high range assay (100 to 5000
ng/mL final DNA concentration) is selected, prepare
2 mL of 1000 ng/mL DNA by adding 2 µL 1000 µg/mL DNA
to 2 ml high range assay solution prepared in step
4.4.
5.2
Check that lamp and filters are installed correctly
according to the TD-700 Fluorometer Operating Manual.
5.3
Turn on fluorometer and allow to warm up for 10 minutes
(600 seconds).
5.4
Set-up parameters and calibrate fluorometer (refer
to your Operating Manual for detailed instructions).
Calibration with more than 1 standard is recommended
(see section 6.).
5.5
Measure the fluorescence of unknown samples by adding
2 µL unknown sample to 2 mL assay solution used for
standards and blank.
5.6
Insert into sample chamber, close lid, and press [*].
6.
Generating a Standard Curve
Generating
a standard curve verifies the linearity of the assay
within a particular concentration range. It is recommended
that you perform this at least once when working with
a new instrument or performing the assay for the first
time. Also, you may want to generate a standard curve
every few weeks as a quality check on the standard,
a reliability check on the instrument, and a consistency
check on technique.
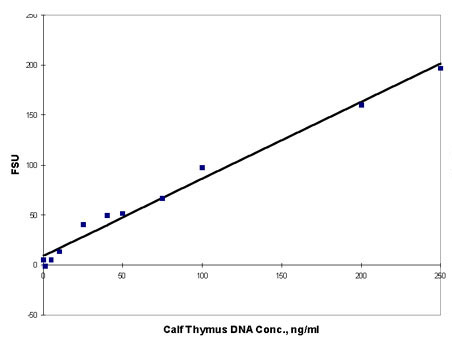
Figure
1. Calf Thymus DNA stained with Hoechst 33258 Dye
dye and fluorescence
measured on Turner BioSystems TD-700 Laboratory Fluorometer.
NOTE:
If the measured values near one end of the curve
deviate consistently from the line, those values represent
a nonlinear region. Sample concentrations should be
adjusted to stay within the linear region of the assay.
|

